Table des matières
You can use the following misc bibtex entry to cite the compute cluster in your posts:
@Misc{HPC_LERIA, title = {High Performance Computing Cluster of LERIA}, year = {2018}, note = {slurm/debian cluster of 27 nodes(700 logical CPU, 2 nvidia GPU tesla k20m, 1 nvidia P100 GPU), 120TB of beegfs scratch storage} }
Presentation of the high performance computing cluster "stargate"
- This wiki page is also yours, do not hesitate to modify it directly or to propose modifications to technique [at] info.univ-angers.fr.
- All cluster users must be on the mailing list calcul-hpc-leria
- To subscribe to this mailing list, simply send an email to sympa@listes.univ-angers.fr with the subject subscribe calcul-hpc-leria Name Surname
Summary
Stargate is the high performance computing cluster of the LERIA computing center. It is a set of 27 computing servers counting 700 CPU cores and 3 GPUs. We also have high performance storage beegfs distributed on 5 storage servers. All these servers are connected to each other via a redundant network with very high speed throughput and very low latency. Services for the functioning of our computing cluster are hosted on the proxmox pool of our computing center. We chose Debian as the operating system for all our servers and slurm for our submission software of job.
Who can use stargate?
In order of priority:
- All members and associate members of the LERIA laboratory,
- The research professors of the University of Angers if they have had the prior authorization of the director of LERIA,
- Visiting researchers if they have the prior authorization of the head chef of the director of LERIA.
- To obtain access to the cluster, simply request the activation of his account by sending an email to technique (at) info.univ-angers.fr
Technical presentation
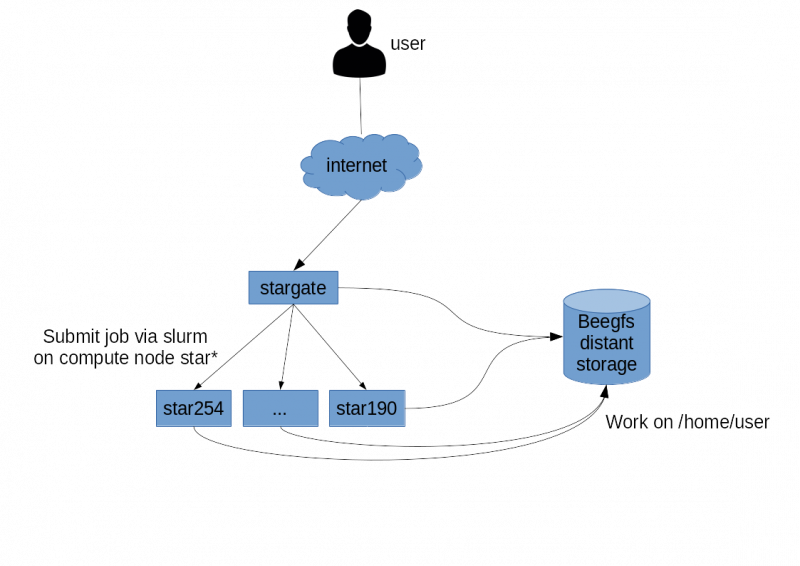
Global architecture
You can also seedata storage
Hardware architecture
| Hostname | Modèle | Nombre de modèle identique | GPU | Nombre de GPU/modèle | CPU | Nombre de CPU/modèle | Nombre de cœurs/CPU | Nombre de threads/CPU | Nombre de threads/modèle | RAM | Espace de stockage local | Interconnection |
| star[254-253] | Dell R720 | 2 | Tesla K20m | 2 | intel-E5-2670 | 2 | 10 | 20 | 40 | 64 Go | 1To | 2*10Gb/s |
| star[246-252] | Dell R620 | 7 | X | 0 | intel-E5-2670 | 2 | 10 | 20 | 40 | 64 Go | 1 To | 2*10Gb/s |
| star[245-244] | Dell R630 | 2 | X | 0 | intel-E5-2695 | 2 | 18 | 36 | 72 | 128 Go | X | 2*10Gb/s |
| star243 | Dell R930 | 1 | X | 0 | intel-E7-4850 | 4 | 16 | 32 | 128 | 1500 Go | 1To | 2*10Gb/s |
| | Dell R730 | 1 | Tesla P100 | 1 | intel-E5-2620 | 2 | 8 | 16 | 32 | 128 Go | 1 To | 2*10Gb/s |
| star[199-195] | Dell R415 | 5 | X | 0 | amd-opteron-6134 | 1 | 8 | 16 | 16 | 32 Go | 1 To | 2*1Gb/s |
| star[194-190] | Dell R415 | 5 | X | 0 | amd-opteron-4184 | 1 | 6 | 12 | 12 | 32 Go | 1 To | 2*1Gb/s |
| star100 | Dell T640 | 1 | RTX 2080 Ti | 4 | intel-xeon-bronze-3106 | 1 | 8 | 16 | 16 | 96 Go | X | 2*10 Gb/s |
| star101 | Dell R740 | 1 | Tesla V100 32 Go | 3 | intel-xeon-server-4208 | 2 | 8 | 16 | 32 | 96 Go | X | 2*10 Gb/s |
Software architecture
The software architecture for the submission of tasks is based on the Slurm tool. Slurm is an open source, fault-tolerant, and highly scalable cluster planning and management system designed for Linux clusters. In the sense of Slurm, the nodes (servers) of calculations are named nodes, and these nodes are grouped in family called partition (which have nothing to do with the notion of partition which segments a peripheral mass storage)
Our cluster has 5 named partitions:
- gpu
- intel-E5-2695
- ram
- amd
- std
Each of these partitions contains nodes.
The compute nodes work with a debian stable operating system. You can find the list of installed software in the List of installed software for high performance computing sections.
Usage policy
- compress your important data
- move your important compressed data to another storage space
- backup your important compressed data
- delete unnecessary and unused data
- You name files and directories should not contain:
- space
- accented characters (é, è, â, …)
- symbols (*, $,%, …)
- punctuation (!,:,; ,,, …)
System administrators reserve the right to rename, compress, delete your files at any time.
There is no backup of your files on the compute cluster, you can lose all your data at any time!
In addition, to avoid uses that could affect other users, a quota of 50 GB is applied to your home directory. Users requiring more space should make an explicit request to technical [at] info.univ-angers.fr. You can also request access to a large capacity storage for a limited time: all data that has been present for more than 40 days in this storage are automatically deleted without the possibility of recovery.
Using the high performance computing cluster
Quick start
Connection to stargate
Please contact technique [at] info.univ-angers.fr to get information about connexion to the compute cluster.
https://www.linode.com/docs/security/authentication/use-public-key-authentication-with-ssh/
Slurm: first tests and documentation
Slurm (Simple Linux Utility for Resource Management) is a task scheduler. Slurm determines where and when the calculations are distributed on the different calculation nodes according to:
- the current load of these computing servers (CPU, Ram, …)
- user history (notion of fairshare, a user using the cluster will take precedence over a user who uses the cluster a lot)
It is strongly advised to read this documentation before going further.
Once connected, you can type the command sinfo which will inform you about the available partitions and their associated nodes:
username_ENT@stargate:$ sinfo PARTITION AVAIL TIMELIMIT NODES STATE NODELIST gpu up 14-00:00:0 2 idle star[242,254] intel-E5-2695 up 14-00:00:0 1 idle star245 amd-opteron-4184 up 14-00:00:0 5 idle star[190-194] std* up 14-00:00:0 6 idle star[190-194,245] ram up 14-00:00:0 1 idle star243 username_ent@stargate:~$
There are two main ways to submit jobs to Slurm:
- Interactive execution, (via the command srun).
- Execution in batch mode (via the command sbatch).
Batch mode execution is presented later in this wiki.
Interactive execution
In order to submit a job to slurm, simply prefix the name of the executable with the command srun.
In order to understand the difference between a process supported by the stargate OS and a process supported by slurm, you can for example type the following two commands:
username_ENT@stargate:~$ hostname stargate username_ENT@stargate:~$ srun hostname star245 username_ENT@stargate:~$
For the first command, the return of hostname gives stargate while the second command srun hostname returns star245. star245 is the name of the machine that has been dynamically designated by slurm to execute the hostname command.
You can also type the commands srun free -h or srun cat /proc/cpu_info to learn more about the nodes of a partition.
Whenever slurm is asked to perform a task, it places it in a thread also called queue. The command squeue allows you to know the list of tasks being processed. This is a bit like the GNU/linux ps command aux or ps -efl but for cluster jobs rather than processes. We can test this command by launching for example on one side srun sleep infinity &. While running this task, the squeue command will give:
username_ENT@stargate:~$ squeue
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
278 intel-E5- sleep username R 0:07 1 star245
It is possible to kill this task via the command scancel with the job identifier as argument:
username_ENT@stargate:~$ scancel 278 username_ENT@stargate:~$ srun: Force Terminated job 332 srun: Job step aborted: Waiting up to 32 seconds for job step to finish. slurmstepd: error: *** STEP 332.0 ON star245 CANCELLED AT 2018-11-27T11:42:06 *** srun: error: star245: task 0: Terminated [1]+ Termine 143 srun sleep infinity username_ENT@stargate:~$
Documentation
To go further, you can watch this series of video presentation and introduction to slurm (in 8 parts):
<html> <center> <iframe width=“500” height=“281” src=“https://www.youtube.com/embed/NH_Fb7X6Db0?list=PLZfwi0jHMBxB-Bd0u1lTT5r0C3RHUPLj-” frameborder=“0” allow=“autoplay; encrypted-media” allowfullscreen></iframe> </center> </html>
You will find here the official slurm documentation.
Hello world !
Compilation
The Stargate machine is not a compute node: it is a node from which you submit your calculations on compute nodes. Stargate is said to be a master node. Therefore, source codes will be compiled on the compute nodes prefixing the compilation with the srun command.
Let's look at the following file named main.cpp:
#include <iostream> int main() { std::cout<<"Hello world!"<<std::endl; return 0; }
It can be compiled using one of the nodes of the partition intel-E5-2695 via the command:
username_ENT@stargate:~$ srun --partition=intel-E5-2695 g++ -Wall main.cpp
username_ENT@stargate:~$ srun g++ -Wall main.cpp
Interactive execution
Finally, we can run this freshly compiled program with
user@stargate:~$ srun -p intel-E5-2695 ./hello
Most of the time, an interactive execution will not interest you, you will prefer and you must use the submission of a job in batch mode. Interactive execution can be interesting for compilation or for debugging.
Batch mode execution
- It's in this mode that a computing cluster is used to execute its processes
- You must write a submission script, this one contains 2 sections:
- The resources you want to use
- Variables for slurm are preceded by #SBATCH
- The commands needed to run the program
Example
#!/bin/bash # hello.slurm #SBATCH --job-name=hello #SBATCH --output=hello.out #SBATCH --error=hello.err #SBATCH --mail-type=end #SBATCH --mail-user=user@univ-angers.fr #SBATCH --nodes=1 #SBATCH --ntasks-per-node=1 #SBATCH --partition=intel-E5-2695 /path/to/hello && sleep 5
user@stargate:~$ sbatch hello.slurm # Job submission user@stargate:~$ squeue # Place and status of jobs in the submission queue user@stargate:~$ cat hello.out # Displays what the standard output would have shown in interactive mode (respectively hello.err for error output)
Very often, we want to run a single program for a set of files or a set of parameters, in which case there are 2 solutions to prioritize:
- use an array job (easy to use, it is the preferred solution).
- use the steps job (more complex to implement).
IMPORTANT: Availability and Resource Management Policy
Slurm is a scheduler. Planning is a difficult and resource-intensive optimization problem. It is much easier for a scheduler to plan jobs if he knows about:
* its duration * the resources to use (CPU, Memory)
In fact, default resources have been defined:
* the duration of a job is 20 minutes * the available memory per CPU is 200 MB
It is quite possible to override these default values with the –mem-per-cpu and –time options. However,
- You should not overstate the resources of your jobs. In fact, slurm works with a fair share concept: if you book resources, whatever you use them or not. In future submissions, slurm will consider that you have actually consumed those resources. Potentially, you could be considered a greedy user and have lower priority than a user who has correctly defined his resources for the same amount of work done.
- If you have a large number of jobs to do, you must use submission by array job .
- If these jobs have long execution times (more than 1 day), you must limit the number of parallel executions in order to not saturate the cluster . We allow users to set this limit, but if there is a problem sharing resources with other users, we will delete jobs that don't respect these conditions .
Limitations
| MaxWallDurationPerJob | MaxJobs | MaxSubmitJobs | FairSharePriority | |
| leria-user | 14 days | 10000 | 99 | |
| guest-user | 7 days | 20 | 50 | 1 |
Disk space quota
See also usage policy and data storage.
By default the quota of disk space is limited to 50GB. You can easily find out which files take up the most space with the command:
user@stargate~ # ncdu
Data storage
You can also see global architecture.
- The compute cluster uses a pool of distributed storage servers beegfs. This beegfs storage is independent of the compute servers. This storage area is naturally accessible in the tree of any compute node under /home/$USER. Since this storage is remote, all read/write in your home is network dependent. Our Beegfs storage and the underlying network are very powerful, but for some heavy processing, you might be better off using local disks from the compute servers. To do this, you can use the /local_working_directory directory of the calculation servers. This directory works in the same way as /tmp except that the data is persistent when the server is restarted.
- If you want to create groups, please send an email to technique.info [at] listes.univ-angers.fr with the name of the group and the associated users.
- As a reminder, by default, the rights of your home are in 755, so anyone can read and execute your data.
Advanced use
Array jobs
You should start by reading the official documentation. This page presents some interesting use case.
If you have a large number of files or parameters to process with a single executable, you must use a array job.
It's easy to implement, just add the –array option to our batch script:
Parametric tests
It's easy to use the array jobs to do parametric tests. That is, use the same executable, possibly on the same file, but by varying a parameter in options of the executable. For that, if the parameters are contiguous or regular, one will use a batch like this one:
#!/bin/bash #SBATCH -J Job_regular_parameter #SBATCH -N 1 #SBATCH --ntasks-per-node=1 #SBATCH -t 10:00:00 #SBATCH --array=0-9 #SBATCH -p intel-E5-2670 #SBATCH -o %A-%a.out #SBATCH -e %A-%a.err #SBATCH --mail-type=end,fail #SBATCH --mail-user=username@univ-angers.fr /path/to/exec --paramExecOption $SLURM_ARRAY_TASK_ID
The –array option can take special syntaxes, for irregular values or for value jumps:
# irregular values 0,3,7,11,35,359 --array=0,3,7,11,35,359 # value jumps of +2: 1, 3, 5 et 7 --array=1-7:2
Multiple instances job
It is common to run a program many times over many instances (benchmark).
Let the following tree:
job_name ├── error ├── instances │ ├── bench1.txt │ ├── bench2.txt │ └── bench3.txt ├── job_name_exec ├── output └── submit_instances_dir.slurm
It is easy to use an array job to execute job_name_exec on all the files to be processed in the instances directory. Just run the following command:
mkdir error output 2>/dev/null || sbatch --job-name=$(basename $PWD) --array=0-$(($(ls -1 instances|wc -l)-1)) submit_instances_dir.slurm
with the following batch file submit_instances_dir.slurm:
#!/bin/bash #SBATCH --mail-type=END,FAIL #SBATCH --mail-user=YOUR-EMAIL #SBATCH -o output/%A-%a #SBATCH -e error/%A-%a #INSTANCES IS ARRAY OF INSTANCE FILE INSTANCES=(instances/*) ./job_name_exec ${INSTANCES[$SLURM_ARRAY_TASK_ID]}
Multiple instances job with multiple executions (Seed number)
Sometimes it is necessary to launch several times the execution on an instance by modifying the seed which makes it possible to generate and reproduct random numbers.
Let the following tree:
job_name ├── error ├── instances │ ├── bench1.txt │ ├── bench2.txt │ └── bench3.txt ├── job_name_exec ├── output ├── submit_instances_dir_with_seed.slurm └── submit.sh
Just run the following command:
./submit.sh
with the following submit.sh file (remember to change the NB_SEED variable):
#!/bin/bash readonly NB_SEED=50 for instance in $ (ls instances) do sbatch --output output/${instance}_%A-%a --error error/${instance}_%A-%a --array 0-${NB_SEED} submit_instances_dir_with_seed.slurm instances/${instance} done exit 0
and the following submit_instances_dir_with_seed.slurm batch:
#!/bin/bash #SBATCH --mail-type = END, FAIL #SBATCH --mail-user = YOUR-EMAIL echo "####### INSTANCE: $ {1}" echo "####### SEED NUMBER: $ {SLURM_ARRAY_TASK_ID}" echo srun echo nameApplication $ {1} $ {SLURM_ARRAY_TASK_ID}
With this method, the variable SLURM_ARRAY_TASK_ID contains the seed. And you submit as many array jobs as there are instances in the instance directory. You can easily find your output which is named like this:
output/instance_name-ID_job-seed_number
Dependencies between job
You can determine dependencies between jobs through the –depend sbatch options:
Example
# job can begin after the specified jobs have started sbatch --depend=after:123_4 my.job #job can begin after the specified jobs have run to completion with an exit code of zero sbatch --depend=afterok:123_4:123_8 my.job2 # job can begin after the specified jobs have terminated sbatch --depend=afterany:123 my.job # job can begin after the specified array_jobs have completely and successfully finish sbatch --depend=afterok:123 my.job
You can also see this page.
Steps jobs
You can use the steps jobs for multiple and varied executions.
The steps jobs:
- allow to split a job into several tasks
- are created by prefixing the program command to be executed by the command Slurm “srun”
- can run sequentially or in parallel
Each step can use n tasks (task) on N compute nodes (-n and -N options of srun). A task has CPU-per-task CPU at its disposal, and there are ntasks allocated by step.
Example
#SBATCH --job-name=nameOfJob #SBATCH --cpus-per-task=1 # Allocation of 1 CPUs per task #SBATCH --ntasks=2 # Tasks number : 2 #SBATCH --mail-type=END # Email notification #SBATCH --mail-user=username@univ-angers.fr # at the end of job. # Step of 2 Tasks srun before.sh # 2 Step in parallel (because &): task1 and task2 run in parallel. There is only one task per Step (option -n1) srun -n1 -N1 /path/to/task1 -threads $SLURM_CPUS_PER_TASK & srun -n1 -N1 /path/to/task2 -threads $SLURM_CPUS_PER_TASK & # We wait for the end of task1 and task2 before running the last step after.sh wait srun after.sh
Steps creation shell bash structure according to the source of the data
From here
Example
# Loop on the elements of an array (here files) : files=('file1' 'file2' 'file3' ...) for f in "${files[@]}"; do # Adapt "-n1" and "-N1" according to your needs srun -n1 -N1 [...] "$f" & done # Loop on the files of a directory: while read f; do # Adapt "-n1" and "-N1" according to your needs srun -n1 -N1 [...] "$f" & done < <(ls "/path/to/files/") # Use "ls -R" or "find" for a recursive file path # Reading line by line of a file: while read line; do # Adapt "-n1" and "-N1" according to your needs srun -n1 -N1 [...] "$line" & done <"/path/to/file"
Use OpenMp ?
Just add the –cpus-per-task option and export the variable OMP_NUM_THREADS
#!/bin/bash # openmp_exec.slurm #SBATCH --job-name=hello #SBATCH --output=openmp_exec.out #SBATCH --error=openmp_exec.err #SBATCH --mail-type=end #SBATCH --mail-user=user@univ-angers.fr #SBATCH --nodes=1 #SBATCH --ntasks-per-node=1 #SBATCH --partition=intel-E5-2695 #SBATCH --cpus-per-task=20 export OMP_NUM_THREADS=20 /path/to/openmp_exec
Specific use
Ssh access of compute nodes
By default, it's impossible to connect ssh directly to the compute nodes. However, if it's justified, we can easily make temporary exceptions. In this case, please make an explicit request to technique [at] info.univ-angers.fr
Users with ssh access must be subscribed to the calcul-hpc-leria-no-slurm-mode@listes.univ-angers.fr list. To subscribe to this mailing list, simply send an email to sympa@listes.univ-angers.fr with the subject: subscribe calcul-hpc-leria-no-slurm-mode Last name First name
Default rule: we don't launch a calculation on a server on which another user's calculation is already running, even if this user doesn't use all the resources. Exception for boinc processes. These processes pause when you perform your calculations.
The htop command lets you know who is calculating with which resources and for how long.
If in doubt, contact the user who calculates directly by email or via the calcul-hpc-leria-no-slurm-mode@listes.univ-angers.fr list.
Cuda
GPU cards are present on star nodes {242,253,254}:
- star242: P100
- star253: 2*k20m
- star254: 2*k20m
Currently, version 9.1 of cuda-sdk-toolkit is installed.
These nodes are currently out of slurm submission lists (although the gpu partition already exists). To be able to use it, thank you to make the explicit request to technique [at] info.univ-angers.fr
RAM node
The leria has a node consisting of 1.5 TB of ram, it's about star243.
This node is accessible by submission via slurm (ram partition). To be able to use it, thank you to make the explicit request to technique [at] info.univ-angers.fr
Cplex
Leria has an academic license for the Cplex software.
The path to the library cplex is the default path /opt/ibm/ILOG/CPLEX_Studio129 (version 12.9)
Conda environments (Python)
The conda activate <env_name> command, activating a conda environnement is unavailable with slurm. You should rather use at the beginning of your script :
source ./anaconda3/bin/activate <env_name>
It may also be necessary to update the environment variables and to initialize conda on the node :
source .bashrc conda init bash
The environment will stay active after your tasks are over. To deactivate the environment, you should use :
source ./anaconda3/bin/deactivate
FAQ
- How to know which are the resources of a partition, example with the partition std:
user@stargate~# scontrol show Partition std
- What means “Some of your processes may have been killed by the cgroup out-of-memory handler” in the standard output of your job ?
You have exceeded the memories limits (–mem-per-cpu parameters)
- How to get an interactive shell prompt in a compute node of your default partition?
user@stargate~# salloc
srun -n1 -N1 --mem-per-cpu=0 --pty --preserve-env --cpu-bind=no --mpi=none $SHELL
- How to get an interactive shell prompt in a specific compute node?
user@stargate~# srun -w NODE_NAME -n1 -N1 --pty bash -i user@NODE_NAME~#
- How can I quote the resources of LERIA in my scientific writings?
You can use the following misc bibtex entry to cite the compute cluster in your posts:
@Misc{HPC_LERIA, title = {High Performance Computing Cluster of LERIA}, year = {2018}, note = {slurm/debian cluster of 27 nodes(700 logical CPU, 2 nvidia GPU tesla k20m, 1 nvidia P100 GPU), 120TB of beegfs scratch storage} }
Error during job submission
- When submitting my jobs, I get the following error message:
srun: error: Unable to allocate resources: Requested node configuration is not available
This probably means that you are trying to use a node without having to specify the partition in which it's located. You must use the -p or –partition option followed by the name of the partition in which the node is located. To have this information, you can do:
user@stargate# scontrol show node NODE_NAME|grep Partitions
Lists of install software for high performance calculating
Via apt-get
- automake
- bison
- boinc-client
- bowtie2
- build-essential
- cmake
- flex
- freeglut3
- freeglut3-dev
- g++
- g++-8
- g++-7
- g++-6
- git
- glibc-doc
- glibc-source
- gnuplot
- libglpk-dev
- libgmp-dev
- liblapack3
- liblapack-dev
- liblas3
- liblas-dev
- libtool
- libopenblas-base
- maven
- nasm
- openjdk-8-jdk-headless
- r-base
- r-base-dev
- regina-rexx
- samtools
- screen
- strace
- subversion
- tmux
- valgrind
- valgrind-dbg
- valgrind-mpi
Via pip
- keras
- scikit-learn
- tenserflow
- tenserflow-gpu # Sur nœuds gpu
GPU node via apt-get
- libglu1-mesa-dev
- libx11-dev
- libxi-dev
- libxmu-dev
- libgl1-mesa-dev
- linux-source
- linux-headers
- linux-image
- nvidia-cuda-toolkit
Software installation
Maybe a program is missing from the list above. In this case, 5 options are available to you:
- Make a request to technique [at] info.univ-angers.fr of the software you want to install
- Make yourself the installation via pip, pip2 or pip3
- Make yourself the installation by compiling the sources in your home_directory
Visualize the load of the high-performance computing cluster
For the links below, you will need to authenticate with your login and password ldap (idem ENT).
Cluster load overview
Details per node
https://grafana.leria.univ-angers.fr/d/000000007/noeuds-du-cluster